REFECT –REcurrent FEatures for Combination Therapy
REFECT is a bioinformatic tool being used to identify genomic and proteomic aberrations developed by the Korkut Lab.
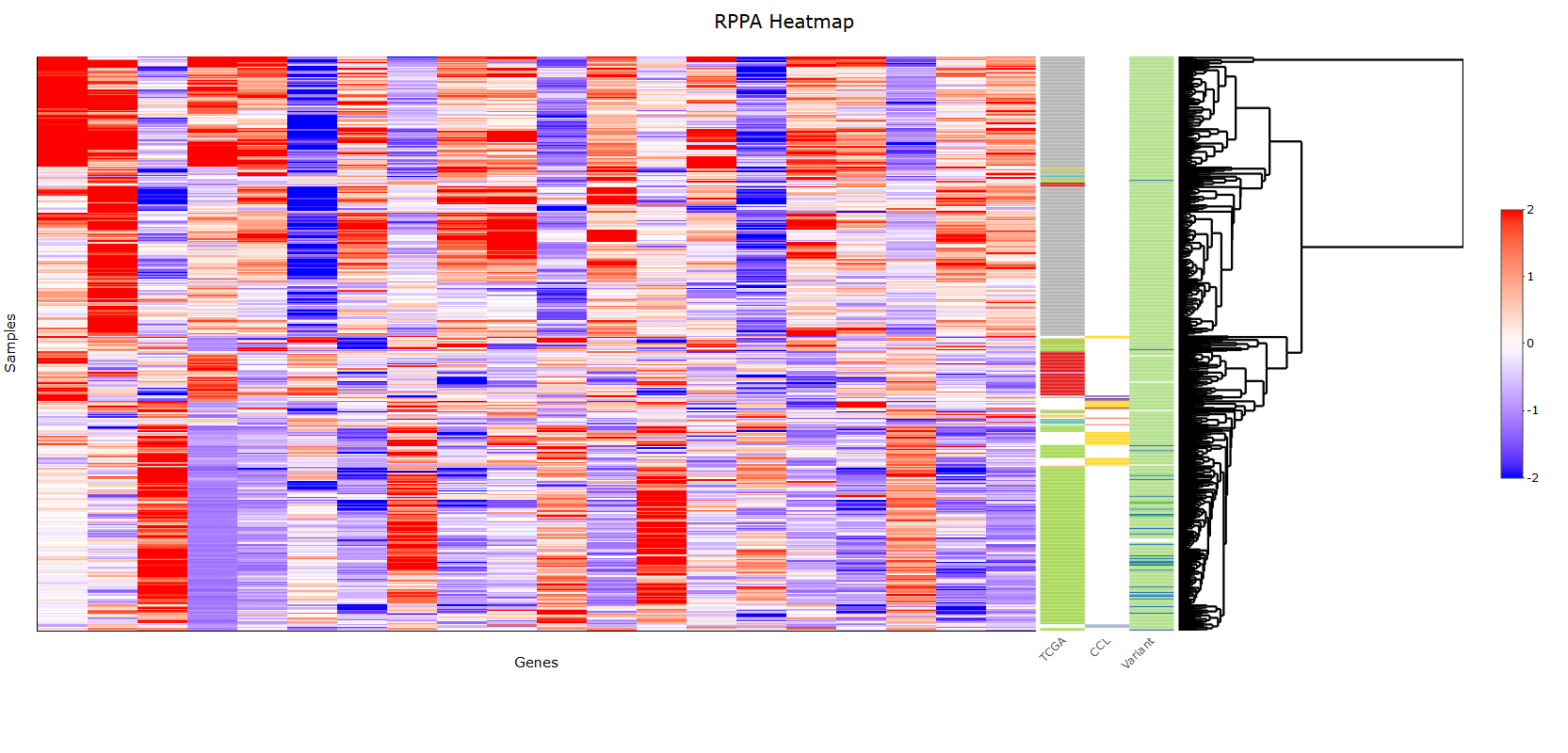
This R-shiny based online resource REFECT algorithm uses proteomic, transciptomic, mutational and copy number data to reveal recurrent, actionable and co-existing alterations in colorectal cancer patients and cell lines.
https://korkutlab.shinyapps.io/refect/
Motivation and Purpose Precision medicine has focused on developing monotherapies based on genomic markers of response, however these therapies have failed to induce durable responses. Multiple studies have demonstrated that rational combination therapies are more effective than monotherapies since they can block multiple oncogenic processes. Therefore, we have established a platform for the detection of recurrent, co-occurring actionable features that are likely oncogenic drivers in order to systematically select and prioritize combination therapies.
Method We began by integrating and analyzing the data from genomically characterized patients (TCGA) and cell line data through z-score based normalization in order to have one integrated data set. To identify a small set of actionable features that discriminate classes of patients and avoid the complexities of high-dimensional data, we implemented a sparse clustering-based feature selection. The algorithm optimizes the dissimilarity matrix between 5 clusters iteratively and uses a LASSO-type penalty for the discriminant feature weights. We designed a multivariate analysis to select actionable co-targets in each sub-cohort that has recurrent characteristics and significantly strong readouts