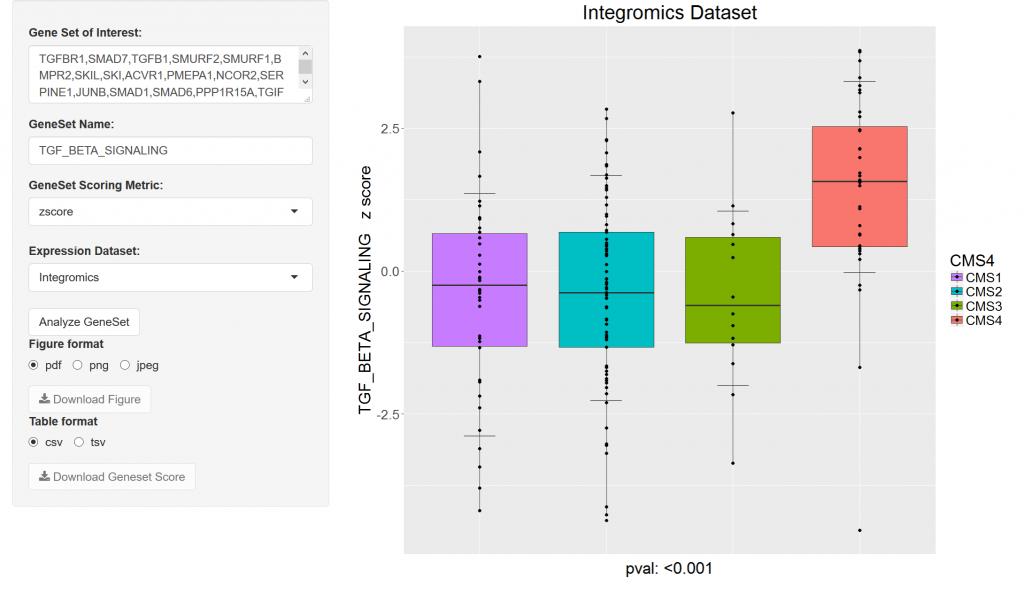
This shiny app is designed to evaluate differences in gene signatures between the CMS groups of single genes. The genes must be entered in using standard nomenclature and separated by a comma. Three datasets are available, including the proprietary internal Integromics dataset, a batch normalized set of Affymetric array data, and the TCGA. Three different methodologies are provided for generating the signatures. Links are provided for downloading the raw data and the image files. Please contact Gani Manyam (gcmanyam@mdanderson.org) if server is unavailable.
 Open Access
Open Access
https://bioinformatics.mdanderson.org/apps/CMS_GeneSetAnalysis/
*This format can be achieved for larger gene sets by first exporting the list to an Excel files with genes listed in a single row, then saving in a CSV format, and then opening with a text editor and copy/pasting the resulting text file.